Drug Discovery
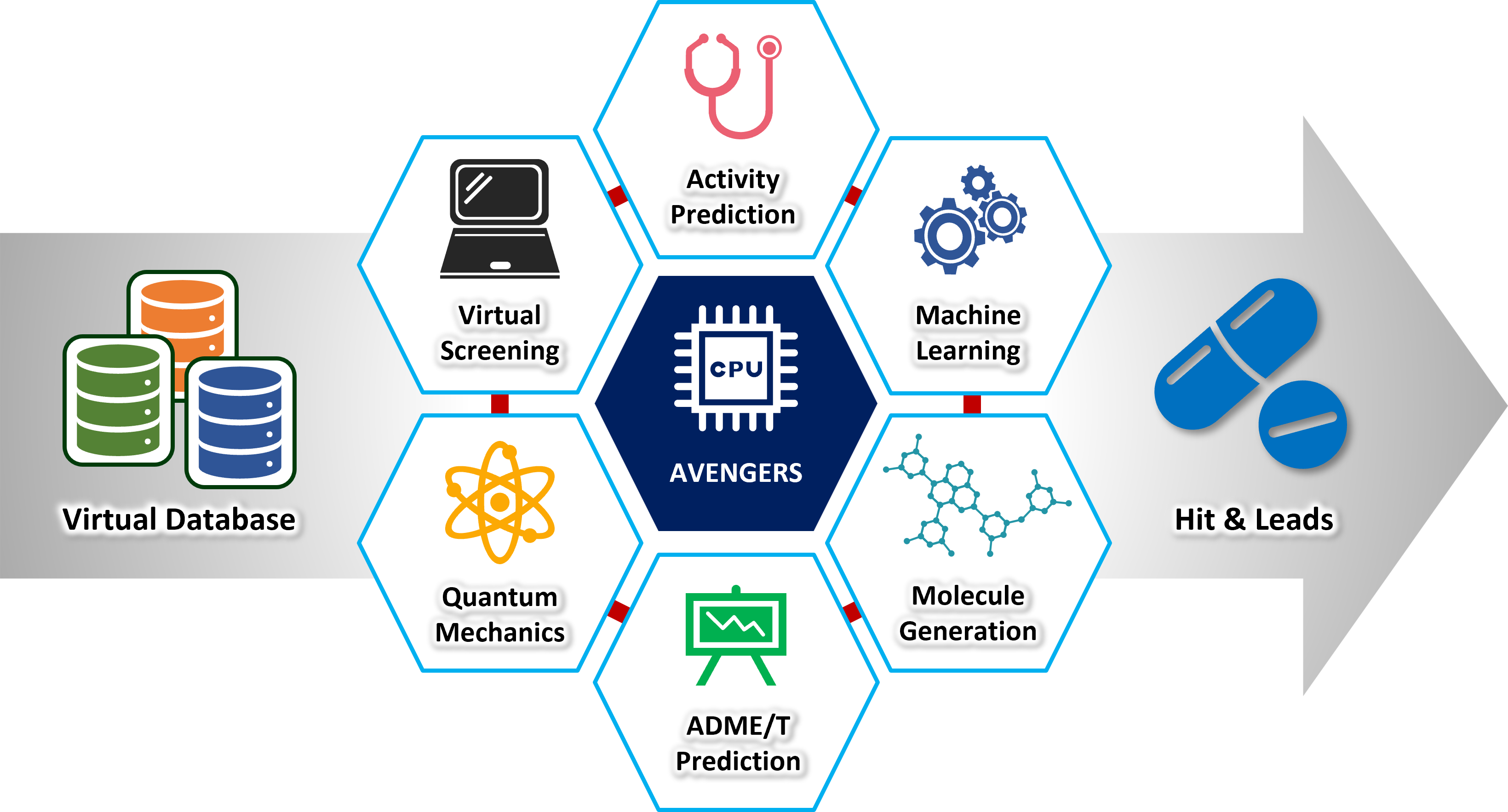
AVENGERS Accelerates Drug Discovery with Precision and Efficiency!
Virtual Screening
Drug Design
It provides a set of compounds by analyzing and selecting potential hit molecules from large databases or even scratch.
Lead Optimization
Drug Design
It optimizes molecules from the first hits with/without structural similarity by quantitative structure-activity relationship and reinforcement learning.
ADME-T Prediction
Drug Design
It predicts ADME-T characteristics of small molecules and suggests new derivatives with better ADME-T profiles than original.